John McDowell
- School of Plant and Environmental Sciences
- College of Agriculture and Life Sciences
Synopsis:
Dr. McDowell uses genetic and genomics tools to understand how pathogens manipulate plants at the molecular level to cause disease. He also seeks to understand how plants recognize pathogens and mount an effective self-defense response. Lastly, he develops new approaches for breeding disease resistant crops (e.g. soybean).
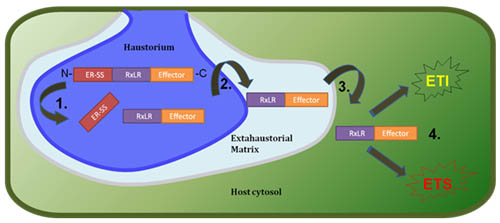
Description:
Pathogens have evolved sophisticated molecular weapons to exploit plants as sources of food and shelter. For example, many pathogens have evolved the capacity to export their own proteins to the interior of plant cells. Once they have gained entry to the interior of plant cells, these pathogen "effector" proteins manipulate specific plant regulatory proteins to make the plant more susceptible to infection. Plants, in turn, have evolved large collections of surveillance proteins that recognize specific pathogen molecules (including some effector proteins) as signals of invasion. This molecular recognition can trigger potent immune responses in the plant, including programmed plant cell suicide at the site of invasion.
My group investigates the molecular interplay and co-evolution between pathogens effector proteins, their targets inside plant cells, and the plant immune surveillance system. Most of our effort is focused on the interaction between the model plant Arabidopsis thaliana and the oomycete pathogen Hyaloperonospora arabidopsidis (downy mildew disease). H. arabidopsidis is a natural pathogen of Arabidopsis, and is related to downy mildew pathogens of crops as well as notorious pathogens in the Phytophthora genus (e.g., late blight of potato). We use molecular genetic and genome-enabled approaches to address the following, interrelated questions:
- How do pathogens manipulate plant cells? Several projects addressing to this question are underway. First, we are co-leading a coalition to sequence the H. arabidopsidis genome. This effort has provided insight into the genomic basis and evolution of "obligate biotrophy", a lifestyle in which pathogens are completely dependent on their hosts for survival. In addition, analysis of the genome has revealed a large collection of so-called "RXLR" effector proteins in H. arabidopsidis. We are currently characterizing the function of some of these genes, including their localization inside plant cells and the plant proteins that are targeted by these effectors. In addition, we are part of a coalition to develop proteins microarrays as a tool for high-throughput functional analysis of RXLR effectors. These projects will provide insight into the molecular mechanisms through which oomycete pathogens cause disease, and may reveal novel plant pathways that play a role in interaction with microbes.
- How do plants evolve new surveillance genes? We are collaborating with Dr. John Jelesko to develop a genetic screening system that models the impact of genetic recombination on the evolution of clustered plant surveillance genes ("R genes"). This project will provide a clearer understanding of how frequently resistance genes recombine, whether the frequency of recombination is elevated in response to stress, and whether new resistance genes can be produced from a single recombination.
- Can fundamental research on plant-pathogen interactions be translated into new strategies for disease control?Diseases caused by plant pathogens are a perennial threat to global food security, causing 100's of billions of dollars in damage on an annual basis. We are part of a coalition to leverage oomycete pathogen genome information for control of soybean diseases, including root/stem rot caused by Phytophthora sojae. Our role in this project is to use information from pathogen genomics to search for novel resistance genes in soybean and related species. These genes can be used in molecular breeding for resistance and we expect that they will be durable against pathogen co-evolution.



